evalne.methods package¶
Submodules¶
evalne.methods.katz module¶
-
class
evalne.methods.katz.Katz(G, beta=0.005)[source]¶ Bases:
objectComputes the exact katz similarity based on paths between nodes in the graph. Shorter paths will contribute more than longer ones. This contribution depends of the damping factor ‘beta’. The exact katz score is computed using the adj matrix of the full graph.
Parameters: - G (graph) – A NetworkX graph or digraph with nodes being consecutive integers starting at 0.
- = float, optional (beta) – The damping factor for the model. Default is 0.005.
Notes
The execution is based on dense matrices, so it may run out of memory.
-
get_params()[source]¶ Returns a dictionary of model parameters.
Returns: params – A dictionary of model parameters and their values. Return type: dict
-
class
evalne.methods.katz.KatzApprox(G, beta=0.005, path_len=3)[source]¶ Bases:
objectComputes the approximated katz similarity based on paths between nodes in the graph. Shorter paths will contribute more than longer ones. This contribution depends of the damping factor ‘beta’. The approximated score is computed using all paths between nodes of length at most ‘path_len’.
Parameters: - G (graph) – A NetworkX graph or digraph.
- beta (float, optional) – The damping factor for the model. Default is 0.005.
- path_len (int, optional) – The maximum path length to consider between each pair of nodes. Default is 3.
Notes
The implementation follows the indication in [1]. It becomes extremely slow for large dense graphs.
References
[1] R. Laishram “Link Prediction in Dynamic Weighted and Directed Social Network using Supervised Learning” Dissertations - ALL. 355. 2015
evalne.methods.similarity module¶
-
evalne.methods.similarity.common_neighbours(G, ebunch=None, neighbourhood='in')[source]¶ Computes the common neighbours similarity between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the common neighbours similarity of nodes ‘u’ and ‘v’ is defined as:
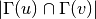
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
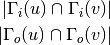
-
evalne.methods.similarity.jaccard_coefficient(G, ebunch=None, neighbourhood='in')[source]¶ Computes the Jaccard coefficient between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the Jaccard coefficient of nodes ‘u’ and ‘v’ is defined as:

For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
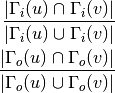
-
evalne.methods.similarity.cosine_similarity(G, ebunch=None, neighbourhood='in')[source]¶ Computes the cosine similarity between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the cosine similarity of nodes ‘u’ and ‘v’ is defined as:
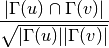
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
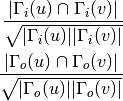
-
evalne.methods.similarity.lhn_index(G, ebunch=None, neighbourhood='in')[source]¶ Computes the Leicht-Holme-Newman index [1]_ between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the Leicht-Holme-Newman index of nodes ‘u’ and ‘v’ is defined as:
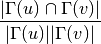
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
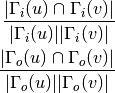
References
[1] Leicht, E. A. and Holme, Petter and Newman, M. E. J. (2006). “Vertex similarity in networks.”, Phys. Rev. E, 73, 10.1103/PhysRevE.73.026120.
-
evalne.methods.similarity.topological_overlap(G, ebunch=None, neighbourhood='in')[source]¶ Computes the topological overlap [2] between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the topological overlap of nodes ‘u’ and ‘v’ is defined as:
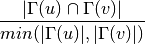
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
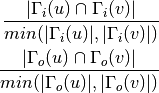
References
[2] Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N., & Barabási, A. L. (2002). “Hierarchical organization of modularity in metabolic networks.” Science, 297(5586), 1551-1555.
-
evalne.methods.similarity.adamic_adar_index(G, ebunch=None, neighbourhood='in')[source]¶ Computes the Adamic-Adar index between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the Adamic-Adar index of nodes ‘u’ and ‘v’ is defined as:
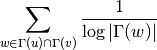
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
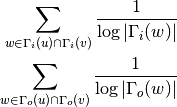
-
evalne.methods.similarity.resource_allocation_index(G, ebunch=None, neighbourhood='in')[source]¶ Computes the resource allocation index between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the resource allocation index of nodes ‘u’ and ‘v’ is defined as:
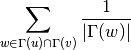
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
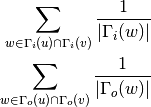
-
evalne.methods.similarity.preferential_attachment(G, ebunch=None, neighbourhood='in')[source]¶ Computes the preferential attachment score between all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs (see Notes for exact definitions).
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.Notes
For undirected graphs the preferential attachment score of nodes ‘u’ and ‘v’ is defined as:
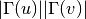
For directed graphs we can consider either the in or the out-neighbourhoods, respectively:
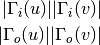
-
evalne.methods.similarity.random_prediction(G, ebunch=None, neighbourhood='in')[source]¶ Returns a float drawn uniformly at random from the interval (0.0, 1.0] for all node pairs in ebunch; or all nodes in G, if ebunch is None. Can be computed for directed and undirected graphs.
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – Not used.
Returns: sim – A list of node-pair similarities in the same order as ebunch.
Return type: list
-
evalne.methods.similarity.all_baselines(G, ebunch=None, neighbourhood='in')[source]¶ Computes a 5-dimensional embedding for all node pairs in ebunch; or all nodes in G, if ebunch is None. Each of the 5 dimensions correspond to the similarity between the nodes as computed by a different function (i.e. CN, JC, AA, RAI and PA). Can be computed for directed and undirected graphs.
Parameters: - G (graph) – A NetworkX graph or digraph.
- ebunch (iterable, optional) – An iterable of node pairs. If None, all edges in G will be used. Default is None.
- neighbourhood (string, optional) – For directed graphs only. Determines if the in or the out-neighbourhood of nodes should be used. Default is ‘in’.
Returns: emb – Column vector containing node-pair embeddings as rows.
Return type: ndarray
Raises: ValueError– If G is directed and neighbourhood is not one of ‘in’ or ‘out’.