evalne.evaluation package¶
Submodules¶
evalne.evaluation.edge_embeddings module¶
-
evalne.evaluation.edge_embeddings.average(X, ebunch)[source]¶ Computes the embedding of each node pair (u, v) in ebunch as the element-wise average of the embeddings of nodes u and v.
Parameters: - X (dict) – A dictionary of {nodeID: embed_vect, nodeID: embed_vect, …}. Dictionary keys are expected to be of type string and values array_like.
- ebunch (iterable) – An iterable of node pairs (u,v) for which the embeddings must be computed.
Returns: emb – A column vector containing node-pair embeddings as rows. In the same order as ebunch.
Return type: ndarray
Notes
Formally, if we use x(u) to denote the embedding corresponding to node u and x(v) to denote the embedding corresponding to node v, and if we use i to refer to the ith position in these vectors, then, the embedding of the pair (u, v) can be computed element-wise as:
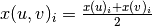 .
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.
.
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.Examples
Simple example of function use and input parameters:
>>> X = {'1': np.array([0, 0, 0, 0]), '2': np.array([2, 2, 2, 2]), '3': np.array([1, 1, -1, -1])} >>> ebunch = ((2, 1), (1, 1), (2, 2), (1, 3), (3, 1), (2, 3), (3, 2)) >>> average(X, ebunch) array([[ 1. , 1. , 1. , 1. ], [ 0. , 0. , 0. , 0. ], [ 2. , 2. , 2. , 2. ], [ 0.5, 0.5, -0.5, -0.5], [ 0.5, 0.5, -0.5, -0.5], [ 1.5, 1.5, 0.5, 0.5], [ 1.5, 1.5, 0.5, 0.5]])
-
evalne.evaluation.edge_embeddings.compute_edge_embeddings(X, ebunch, method='hadamard')[source]¶ Computes the embedding of each node pair (u, v) in ebunch as an element-wise operation on the embeddings of the end nodes u and v. The operator used is determined by the method parameter.
Parameters: - X (dict) – A dictionary of {nodeID: embed_vect, nodeID: embed_vect, …}. Dictionary keys are expected to be of type string and values array_like.
- ebunch (iterable) – An iterable of node pairs (u,v) for which the embeddings must be computed.
- method (string, optional) – The operator to be used for computing the node-pair embeddings. Options are: average, hadamard, weighted_l1 or weighted_l2. Default is hadamard.
Returns: emb – A column vector containing node-pair embeddings as rows. In the same order as ebunch.
Return type: ndarray
Examples
Simple example of function use and input parameters:
>>> X = {'1': np.array([0, 0, 0, 0]), '2': np.array([2, 2, 2, 2]), '3': np.array([1, 1, -1, -1])} >>> ebunch = ((2, 1), (1, 1), (2, 2), (1, 3), (3, 1), (2, 3), (3, 2)) >>> compute_edge_embeddings(X, ebunch, 'average') array([[ 1. , 1. , 1. , 1. ], [ 0. , 0. , 0. , 0. ], [ 2. , 2. , 2. , 2. ], [ 0.5, 0.5, -0.5, -0.5], [ 0.5, 0.5, -0.5, -0.5], [ 1.5, 1.5, 0.5, 0.5], [ 1.5, 1.5, 0.5, 0.5]])
-
evalne.evaluation.edge_embeddings.hadamard(X, ebunch)[source]¶ Computes the embedding of each node pair (u, v) in ebunch as the element-wise product between the embeddings of nodes u and v.
Parameters: - X (dict) – A dictionary of {nodeID: embed_vect, nodeID: embed_vect, …}. Dictionary keys are expected to be of type string and values array_like.
- ebunch (iterable) – An iterable of node pairs (u,v) for which the embeddings must be computed.
Returns: emb – A column vector containing node-pair embeddings as rows. In the same order as ebunch.
Return type: ndarray
Notes
Formally, if we use x(u) to denote the embedding corresponding to node u and x(v) to denote the embedding corresponding to node v, and if we use i to refer to the ith position in these vectors, then, the embedding of the pair (u, v) can be computed element-wise as:
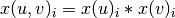 .
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.
.
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.Examples
Simple example of function use and input parameters:
>>> X = {'1': np.array([0, 0, 0, 0]), '2': np.array([2, 2, 2, 2]), '3': np.array([1, 1, -1, -1])} >>> ebunch = ((2, 1), (1, 1), (2, 2), (1, 3), (3, 1), (2, 3), (3, 2)) >>> hadamard(X, ebunch) array([[ 0., 0., 0., 0.], [ 0., 0., 0., 0.], [ 4., 4., 4., 4.], [ 0., 0., 0., 0.], [ 0., 0., 0., 0.], [ 2., 2., -2., -2.], [ 2., 2., -2., -2.]])
-
evalne.evaluation.edge_embeddings.weighted_l1(X, ebunch)[source]¶ Computes the embedding of each node pair (u, v) in ebunch as the element-wise weighted L1 distance between the embeddings of nodes u and v.
Parameters: - X (dict) – A dictionary of {nodeID: embed_vect, nodeID: embed_vect, …}. Dictionary keys are expected to be of type string and values array_like.
- ebunch (iterable) – An iterable of node pairs (u,v) for which the embeddings must be computed.
Returns: emb – A column vector containing node-pair embeddings as rows. In the same order as ebunch.
Return type: ndarray
Notes
Formally, if we use x(u) to denote the embedding corresponding to node u and x(v) to denote the embedding corresponding to node v, and if we use i to refer to the ith position in these vectors, then, the embedding of the pair (u, v) can be computed element-wise as:
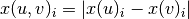 .
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.
.
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.Examples
Simple example of function use and input parameters:
>>> X = {'1': np.array([0, 0, 0, 0]), '2': np.array([2, 2, 2, 2]), '3': np.array([1, 1, -1, -1])} >>> ebunch = ((2, 1), (1, 1), (2, 2), (1, 3), (3, 1), (2, 3), (3, 2)) >>> weighted_l1(X, ebunch) array([[2., 2., 2., 2.], [0., 0., 0., 0.], [0., 0., 0., 0.], [1., 1., 1., 1.], [1., 1., 1., 1.], [1., 1., 3., 3.], [1., 1., 3., 3.]])
-
evalne.evaluation.edge_embeddings.weighted_l2(X, ebunch)[source]¶ Computes the embedding of each node pair (u, v) in ebunch as the element-wise weighted L2 distance between the embeddings of nodes u and v.
Parameters: - X (dict) – A dictionary of {nodeID: embed_vect, nodeID: embed_vect, …}. Dictionary keys are expected to be of type string and values array_like.
- ebunch (iterable) – An iterable of node pairs (u,v) for which the embeddings must be computed.
Returns: emb – A column vector containing node-pair embeddings as rows. In the same order as ebunch.
Return type: ndarray
Notes
Formally, if we use x(u) to denote the embedding corresponding to node u and x(v) to denote the embedding corresponding to node v, and if we use i to refer to the ith position in these vectors, then, the embedding of the pair (u, v) can be computed element-wise as:
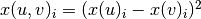 .
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.
.
Also note that all nodeID’s in ebunch must exist in X, otherwise, the method will fail.Examples
Simple example of function use and input parameters:
>>> X = {'1': np.array([0, 0, 0, 0]), '2': np.array([2, 2, 2, 2]), '3': np.array([1, 1, -1, -1])} >>> ebunch = ((2, 1), (1, 1), (2, 2), (1, 3), (3, 1), (2, 3), (3, 2)) >>> weighted_l2(X, ebunch) array([[4., 4., 4., 4.], [0., 0., 0., 0.], [0., 0., 0., 0.], [1., 1., 1., 1.], [1., 1., 1., 1.], [1., 1., 9., 9.], [1., 1., 9., 9.]])
evalne.evaluation.evaluator module¶
-
class
evalne.evaluation.evaluator.LPEvaluator(traintest_split, trainvalid_split=None, dim=128, lp_model=LogisticRegressionCV(cv=5, scoring='roc_auc'))[source]¶ Bases:
objectClass designed to simplify the evaluation of embedding methods for link prediction tasks.
Parameters: - traintest_split (LPEvalSplit) – An object containing the train graph (a subgraph of the full network that spans all nodes) and a set of train edges and non-edges. Test edges are optional. If not provided only train results will be generated.
- trainvalid_split (LPEvalSplit, optional) – An object containing the validation graph (a subgraph of the training network that spans all nodes) and a set of train and valid edges and non-edges. If not provided a split with the same parameters as the train one, but with train_frac=0.9, will be computed. Default is None.
- dim (int, optional) – Embedding dimensionality. Default is 128.
- lp_model (Sklearn binary classifier, optional) – The binary classifier to use for prediction. Default is logistic regression with 5 fold cross validation: LogisticRegressionCV(Cs=10, cv=5, penalty=’l2’, scoring=’roc_auc’, solver=’lbfgs’, max_iter=100))
Notes
In link prediction the aim is to predict, given a set of node pairs, if they should be connected or not. This is generally solved as a binary classification task. For training the binary classifier, we sample a set of edges as well as a set of unconnected node pairs. We then compute the node-pair embeddings of this training data. We use the node-pair embeddings together with the corresponding labels (0 for non-edges and 1 for edges) to train the classifier. Finally, the performance is evaluated on the test data (the remaining edges not used in training plus another set of randomly selected non-edges).
Examples
Instantiating an LPEvaluator without a specific train/validation split (this split will be computed automatically if parameter tuning for any method is required):
>>> from evalne.evaluation.evaluator import LPEvaluator >>> from evalne.evaluation.split import LPEvalSplit >>> from evalne.utils import preprocess as pp >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Create the required train/test split >>> traintest_split = LPEvalSplit() >>> _ = traintest_split.compute_splits(G) >>> # Initialize the LPEvaluator >>> nee = LPEvaluator(traintest_split)
Instantiating an LPEvaluator with a specific train/validation split (allows the user to specify any parameters for the train/validation split):
>>> from evalne.evaluation.evaluator import LPEvaluator >>> from evalne.evaluation.split import LPEvalSplit >>> from evalne.utils import preprocess as pp >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Create the required train/test split >>> traintest_split = LPEvalSplit() >>> _ = traintest_split.compute_splits(G) >>> # Create the train/validation split from the train data computed in the trintest_split >>> # The graph used to initialize this split must, thus, be the train graph from the traintest_split >>> trainvalid_split = EvalSplit() >>> _ = trainvalid_split.compute_splits(traintest_split.TG) >>> # Initialize the LPEvaluator >>> nee = LPEvaluator(traintest_split, trainvalid_split)
-
compute_ee(data_split, X, edge_embed_method)[source]¶ Computes node-pair embeddings using the given node embeddings dictionary and node-pair embedding method. If data_split.test_edges is None, te_edge_embeds will be None.
Parameters: - data_split (a subclass of BaseEvalSplit) – A subclass of BaseEvalSplit object that encapsulates the train/test or train/validation data.
- X (dict) – A dictionary where keys are nodes in the graph and values are the node embeddings. The keys are of type string and the values of type array.
- edge_embed_method (string) – A string indicating the method used to compute node-pair embeddings from node embeddings. The accepted values are any of the function names in evalne.evaluation.edge_embeddings.
Returns: - tr_edge_embeds (matrix) – A Numpy matrix containing the train node-pair embeddings.
- te_edge_embeds (matrix) – A Numpy matrix containing the test node-pair embeddings. Returns None if data_split.test_edges is None.
Raises: AttributeError– If the node-pair embedding operator selected is not valid.
-
compute_pred(data_split, tr_edge_embeds, te_edge_embeds=None)[source]¶ Computes predictions from the given node-pair embeddings. Trains an LP model with the train node-pair embeddings and performs predictions for train and test node-pair embeddings. If te_edge_embeds is None test_pred will be None.
Parameters: - data_split (a subclass of BaseEvalSplit) – A subclass of BaseEvalSplit object that encapsulates the train/test or train/validation data.
- tr_edge_embeds (matrix) – A Numpy matrix containing the train node-pair embeddings.
- te_edge_embeds (matrix, optional) – A Numpy matrix containing the test node-pair embeddings. Default is None.
Returns: - train_pred (array) – The predictions for the train data.
- test_pred (array) – The predictions for the test data. Returns None if te_edge_embeds is None.
-
compute_results(data_split, method_name, train_pred, test_pred=None, label_binarizer=LogisticRegression(solver='liblinear'), params=None)[source]¶ Generates results from the given predictions and returns them. If test_pred is not provided, the Results object will only contain the train scores.
Parameters: - data_split (a subclass of BaseEvalSplit) – A subclass of BaseEvalSplit object that encapsulates the train/test or train/validation data.
- method_name (string) – A string indicating the name of the method for which the results will be created.
- train_pred – The predictions for the train data.
- test_pred (array_like, optional) – The predictions for the test data. Default is None.
- label_binarizer (string or Sklearn binary classifier, optional) – If the predictions returned by the model are not binary, this parameter indicates how these binary predictions should be computed in order to be able to provide metrics such as the confusion matrix. Any Sklear binary classifier can be used or the keyword ‘median’ which will used the prediction medians as binarization thresholds. Default is LogisticRegression(solver=’liblinear’)
- params (dict, optional) – A dictionary of parameters and values to be added to the results class. Default is None.
Returns: results – The evaluation results.
Return type:
-
evaluate_baseline(method, neighbourhood='in', timeout=None)[source]¶ Evaluates the baseline method requested. Evaluation output is returned as a Results object. For Katz neighbourhood=`in` and neighbourhood=`out` will return the same results corresponding to neighbourhood=`in`. Execution time is contained in the results object. If the train/test split object used to initialize the evaluator does not contain test edges, the results object will only contain train results.
Parameters: - method (string) – A string indicating the name of any baseline from evalne.methods to evaluate.
- neighbourhood (string, optional) – A string indicating the ‘in’ or ‘out’ neighbourhood to be used for directed graphs. Default is ‘in’.
- timeout (float or None) – A float indicating the maximum amount of time (in seconds) the evaluation can run for. If None, the evaluation is allowed to continue until completion. Default is None.
Returns: results – The evaluation results as a Results object.
Return type: Raises: TimeoutExpired– If the execution does not finish within the allocated time.TypeError– If the Katz method call is incorrect.ValueError– If the heuristic selected does not exist.
See also
evalne.utils.util.run_function()- The low level function used to run a baseline with given timeout.
Examples
Evaluating the common neighbours heuristic with default parameters. We assume an evaluator (nee) has already been instantiated (see class examples):
>>> result = nee.evaluate_baseline(method='common_neighbours') >>> # Print the results >>> result.pretty_print() Method: common_neighbours Parameters: [('split_id', 0), ('dim', 128), ('eval_time', 0.06909489631652832), ('neighbourhood', 'in'), ('split_alg', 'spanning_tree'), ('fe_ratio', 1.0), ('owa', True), ('nw_name', 'test'), ('train_frac', 0.510061919504644)] Test scores: tn = 1124 [...]
Evaluating katz with beta=0.05 and timeout 60 seconds. We assume an evaluator (nee) has already been instantiated (see class examples):
>>> result = nee.evaluate_baseline(method='katz 0.05', timeout=60) >>> # Print the results >>> result.pretty_print() Method: katz 0.05 Parameters: [('split_id', 0), ('dim', 128), ('eval_time', 0.11670708656311035), ('neighbourhood', 'in'), ('split_alg', 'spanning_tree'), ('fe_ratio', 1.0), ('owa', True), ('nw_name', 'test'), ('train_frac', 0.510061919504644)] Test scores: tn = 1266 [...]
-
evaluate_cmd(method_name, method_type, command, edge_embedding_methods, input_delim, output_delim, tune_params=None, maximize='auroc', write_weights=False, write_dir=False, timeout=None, verbose=True)[source]¶ Evaluates an embedding method and tunes its parameters from the method’s command line call string. This function can evaluate node embedding, node-pair embedding or end to end predictors.
Parameters: - method_name (string) – A string indicating the name of the method to be evaluated.
- method_type (string) – A string indicating the type of embedding method (i.e. ne, ee, e2e). NE methods are expected to return embeddings, one per graph node, as either dict or matrix sorted by nodeID. EE methods are expected to return node-pair emb. as [num_edges x embed_dim] matrix in same order as input. E2E methods are expected to return predictions as a vector in the same order as the input edgelist.
- command (string) – A string containing the call to the method as it would be written in the command line. For ‘ne’ methods placeholders (i.e. {}) need to be provided for the parameters: input network file, output file and embedding dimensionality, precisely IN THIS ORDER. For ‘ee’ methods with parameters: input network file, input train edgelist, input test edgelist, output train embeddings, output test embeddings and embedding dimensionality, 6 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER. For methods with parameters: input network file, input edgelist, output embeddings, and embedding dimensionality, 4 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER. For ‘e2e’ methods with parameters: input network file, input train edgelist, input test edgelist, output train predictions, output test predictions and embedding dimensionality, 6 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER. For methods with parameters: input network file, input edgelist, output predictions, and embedding dimensionality, 4 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER.
- edge_embedding_methods (array-like) – A list of methods used to compute node-pair embeddings from the node embeddings output by NE models. The accepted values are the function names in evalne.evaluation.edge_embeddings. When evaluating ‘ee’ or ‘e2e’ methods, this parameter is ignored.
- input_delim (string) – The delimiter expected by the method as input (edgelist).
- output_delim (string) – The delimiter provided by the method in the output.
- tune_params (string, optional) – A string containing all the parameters to be tuned and their values. Default is None.
- maximize (string, optional) – The score to maximize while performing parameter tuning. Default is ‘auroc’.
- write_weights (bool, optional) – If True the train graph passed to the embedding methods will be stored as weighted edgelist (e.g. triplets src, dst, weight) otherwise as normal edgelist. If the graph edges have no weight attribute and this parameter is set to True, a weight of 1 will be assigned to each edge. Default is False.
- write_dir (bool, optional) – This option is only relevant for undirected graphs. If False, the train graph will be stored with a single direction of the edges. If True, both directions of edges will be stored. Default is False.
- timeout (float or None, optional) – A float indicating the maximum amount of time (in seconds) the evaluation can run for. If None, the evaluation is allowed to continue until completion. Default is None.
- verbose (bool, optional) – A parameter to control the amount of screen output. Default is True.
Returns: results – Returns the evaluation results as a Results object.
Return type: Raises: TimeoutExpired– If the execution does not finish within the allocated time.IOError– If the method call does not succeed.ValueError– If the method type is unknown. If for a method all parameter combinations fail to provide results.
See also
evalne.utils.util.run()- The low level function used to run a cmd call with given timeout.
Examples
Evaluating the OpenNE implementation of node2vec without parameter tuning and with ‘average’ and ‘hadamard’ as node-pair embedding operators. We assume the method is installed in a virtual environment and that an evaluator (nee) has already been instantiated (see class examples):
>>> # Prepare the cmd command for running the method. If running on a python console full paths are required >>> cmd = '../OpenNE-master/venv/bin/python -m openne --method node2vec ' ... '--graph-format edgelist --input {} --output {} --representation-size {}' >>> # Call the evaluation >>> result = nee.evaluate_cmd(method_name='Node2vec', method_type='ne', command=cmd, ... edge_embedding_methods=['average', 'hadamard'], input_delim=' ', output_delim=' ') Running command... [...] >>> # Print the results >>> result.pretty_print() Method: Node2vec Parameters: [('split_id', 0), ('dim', 128), ('owa', True), ('nw_name', 'test'), ('train_frac', 0.51), ('split_alg', 'spanning_tree'), ('eval_time', 24.329686164855957), ('edge_embed_method', 'average'), ('fe_ratio', 1.0)] Test scores: tn = 913 [...]
Evaluating the metapath2vec c++ implementation with parameter tuning and with ‘average’ node-pair embedding operator. We assume the method is installed and that an evaluator (nee) has already been instantiated (see class examples):
>>> # Prepare the cmd command for running the method. If running on a python console full paths are required >>> cmd = '../../methods/metapath2vec/metapath2vec -min-count 1 -iter 20 ' ... '-samples 100 -train {} -output {} -size {}' >>> # Call the evaluation >>> result = nee.evaluate_cmd(method_name='Metapath2vec', method_type='ne', command=cmd, ... edge_embedding_methods=['average'], input_delim=' ', output_delim=' ') Running command... [...] >>> # Print the results >>> result.pretty_print() Method: Metapath2vec Parameters: [('split_id', 0), ('dim', 128), ('owa', True), ('nw_name', 'test'), ('train_frac', 0.51), ('split_alg', 'spanning_tree'), ('eval_time', 1.9907279014587402), ('edge_embed_method', 'average'), ('fe_ratio', 1.0)] Test scores: tn = 919 [...]
-
evaluate_ne(data_split, X, method, edge_embed_method, label_binarizer=LogisticRegression(solver='liblinear'), params=None)[source]¶ Runs the complete pipeline, from node embeddings to node-pair embeddings and returns the prediction results. If data_split.test_edges is None, the Results object will only contain train Scores.
Parameters: - data_split (a subclass of BaseEvalSplit) – A subclass of BaseEvalSplit object that encapsulates the train/test or train/validation data.
- X (dict) – A dictionary where keys are nodes in the graph and values are the node embeddings. The keys are of type string and the values of type array.
- method (string) – A string indicating the name of the method to be evaluated.
- edge_embed_method (string) – A string indicating the method used to compute node-pair embeddings from node embeddings. The accepted values are any of the function names in evalne.evaluation.edge_embeddings.
- label_binarizer (string or Sklearn binary classifier, optional) – If the predictions returned by the model are not binary, this parameter indicates how these binary predictions should be computed in order to be able to provide metrics such as the confusion matrix. Any Sklear binary classifier can be used or the keyword ‘median’ which will used the prediction medians as binarization thresholds. Default is LogisticRegression(solver=’liblinear’).
- params (dict, optional) – A dictionary of parameters and values to be added to the results class. Default is None.
Returns: results – A results object.
Return type:
-
class
evalne.evaluation.evaluator.NCEvaluator(G, labels, nw_name, num_shuffles, traintest_fracs, trainvalid_frac, dim=128, nc_model=LogisticRegressionCV(cv=3, multi_class='ovr'))[source]¶ Bases:
objectClass designed to simplify the evaluation of embedding methods for node classification tasks. The input graphs is assumed to be the entire network. Parameter tuning is performed directly on this complete graph using a train/valid node split of specified size.
Parameters: - G (nx.Graph) – The full graph for which to run the evaluation.
- labels (ndarray) – A numpy array containing nodeIDs as first columns and labels as second column.
- nw_name (string) – A string indicating the name of the network. For result logging purposes.
- num_shuffles (int) – The number of times to repeat the evaluation with different train and test node sets.
- traintest_fracs (array-like) – The fraction of all nodes to use for training.
- trainvalid_frac (float) – The fraction of all training nodes to use for actual model training (the rest are used for validation).
- dim (int, optional) – Embedding dimensionality. Default is 128.
- nc_model (Sklearn binary classifier, optional) – The classifier to use for prediction. Default is logistic regression with 3 fold cross validation: LogisticRegressionCV(Cs=10, cv=3, penalty=’l2’, multi_class=’ovr’)
Notes
In node multi-label classification the aim is to predict the label associated with each graph node. We start the evaluation of this task by computing the embeddings for each node in the graph. Then, we train a classifier with with a subset of these embeddings (the training nodes) and their corresponding labels. Performance is evaluate on a holdout set. For robustness, the performance is generally averaged over multiple executions over different shuffles of the data (different train and test sets). The num_shuffles attribute controls the number of shuffles that will be generated.
Examples
Instantiating an NCEvaluator with default parameters:
>>> from evalne.evaluation.evaluator import NCEvaluator >>> from evalne.utils import preprocess as pp >>> import numpy as np >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Generate some random node labels >>> labels = np.random.choice([1,2,3,4,5], size=len(G.nodes)) >>> # Create pairs of (nodeID, label) and make them a column vector >>> nl_pairs = np.vstack((range(len(G.nodes)), labels)).T >>> # For NC we do not need to create a train test edge split, we can initialize the evaluator directly >>> nee = NCEvaluator(G, labels=nl_pairs, nw_name='test_network', num_shuffles=5, traintest_fracs=[0.8, 0.5], ... trainvalid_frac=0.5)
-
compute_pred(X_train, y_train, X_test=None)[source]¶ Computes predictions from the given embeddings. Trains a NC model with the train node-pair embeddings and performs predictions for train and test embeddings. If te_edge_embeds is None test_pred will be None.
Parameters: - X_train (ndarray) – An array containing the train embeddings.
- y_train (ndarray) – An array containing the train labels.
- X_test (ndarray, optional) – An array containing the test embeddings.
Returns: - train_pred (ndarray) – The label predictions for the train data.
- test_pred (ndarray) – The label predictions for the test data. Returns None if X_test is None.
-
compute_results(method_name, train_pred, train_labels, test_pred=None, test_labels=None, params=None)[source]¶ Generates results from the given predictions and returns them. If test_pred is not provided, the Results object will only contain the train scores.
Parameters: - method_name (string) – A string indicating the name of the method for which the results will be created.
- train_pred (ndarray) – The predictions for the train data.
- test_pred (ndarray, optional) – The predictions for the test data. Default is None.
- params (dict, optional) – A dictionary of parameters and values to be added to the results class. Default is None.
Returns: results – The evaluation results.
Return type:
-
evaluate_cmd(method_name, command, input_delim, output_delim, tune_params=None, maximize='f1_micro', write_weights=False, write_dir=False, timeout=None, verbose=True)[source]¶ Evaluates an embedding method and tunes its parameters from the method’s command line call string. Currently, this function can only evaluate node embedding methods.
Parameters: - method_name (string) – A string indicating the name of the method to be evaluated.
- command (string) – A string containing the call to the method as it would be written in the command line. For ‘ne’ methods placeholders (i.e. {}) need to be provided for the parameters: input network file, output file and embedding dimensionality, precisely IN THIS ORDER.
- input_delim (string) – The delimiter expected by the method as input (edgelist).
- output_delim (string) – The delimiter provided by the method in the output.
- tune_params (string, optional) – A string containing all the parameters to be tuned and their values. Default is None.
- maximize (string, optional) – The score to maximize while performing parameter tuning. Default is ‘f1_micro’.
- write_weights (bool, optional) – If True the train graph passed to the embedding methods will be stored as weighted edgelist (e.g. triplets src, dst, weight) otherwise as normal edgelist. If the graph edges have no weight attribute and this parameter is set to True, a weight of 1 will be assigned to each edge. Default is False.
- write_dir (bool, optional) – This option is only relevant for undirected graphs. If False, the train graph will be stored with a single direction of the edges. If True, both directions of edges will be stored. Default is False.
- timeout (float or None) – A float indicating the maximum amount of time (in seconds) the evaluation can run for. If None, the evaluation is allowed to continue until completion. Default is None.
- verbose (bool, optional) – A parameter to control the amount of screen output. Default is True.
Returns: results – Returns the evaluation results as a list of Results objects (one for each traintest_frac requested and each shuffle). The length of the list returned will thus be num_shuffles * len(traintest_fracs).
Return type: list of Results
Raises: TimeoutExpired– If the execution does not finish within the allocated time.IOError– If the method call does not succeed.
See also
evalne.utils.util.run()- The low level function used to run a cmd call with given timeout.
Examples
Evaluating the OpenNE implementation of node2vec without parameter tuning. We assume the method is installed in a virtual environment and that an evaluator (nee) has already been instantiated (see class examples):
>>> # Prepare the cmd command for running the method. If running on a python console full paths are required >>> cmd = '../OpenNE-master/venv/bin/python -m openne --method node2vec ' ... '--graph-format edgelist --input {} --output {} --representation-size {}' >>> # Call the evaluation >>> result = nee.evaluate_cmd(method_name='Node2vec', command=cmd, input_delim=' ', output_delim=' ') Running command... [...] >>> # Check the results of the first data shuffle of traintest_frac=0.8 >>> result[0].pretty_print() Method: Node2vec_0.8 Parameters: [('dim', 128), ('nw_name', 'test_network'), ('eval_time', 33.22737193107605)] Test scores: f1_micro = 0.177304964539 f1_macro = 0.0975922953451 f1_weighted = 0.107965347267 >>> # Check the results of the first data shuffle of traintest_frac=0.5 >>> result[5].pretty_print() Method: Node2vec_0.5 Parameters: [('dim', 128), ('nw_name', 'test_network'), ('eval_time', 33.22737193107605)] Test scores: f1_micro = 0.173295454545 f1_macro = 0.0590799031477 f1_weighted = 0.0511913933524
Evaluating the metapath2vec c++ implementation without parameter tuning. We assume the method is installed and that an evaluator (nee) has already been instantiated (see class examples):
>>> # Prepare the cmd command for running the method. If running on a python console full paths are required >>> cmd = '../../methods/metapath2vec/metapath2vec -min-count 1 -iter 20 ' ... '-samples 100 -train {} -output {} -size {}' >>> # Call the evaluation >>> result = nee.evaluate_cmd(method_name='Metapath2vec', command=cmd, input_delim=' ', output_delim=' ') Running command... [...] >>> # Check the results of the second data shuffle of traintest_frac=0.8 >>> result.pretty_print() Method: Metapath2vec_0.8 Parameters: [('dim', 128), ('nw_name', 'test_network'), ('eval_time', 23.914228916168213)] Test scores: f1_micro = 0.205673758865 f1_macro = 0.0711656441718 f1_weighted = 0.0807553409041 >>> # Check the results of the second data shuffle of traintest_frac=0.5 >>> result.pretty_print() Method: Metapath2vec_0.5 Parameters: [('dim', 128), ('nw_name', 'test_network'), ('eval_time', 23.914228916168213)] Test scores: f1_micro = 0.215909090909 f1_macro = 0.0710280373832 f1_weighted = 0.0766779949023
-
evaluate_ne(X, method_name, params=None)[source]¶ Runs the NC evaluation pipeline. For each ‘node_frac’ trains a nc_model and uses it to compute predictions which are then returned as a results object. If data_split.test_edges is None, the Results object will only contain train Scores.
Parameters: - X (dict) – A dictionary where keys are nodes in the graph and values are the node embeddings. The keys are of type string and the values of type array.
- method_name (string) – A string indicating the name of the method to be evaluated.
- params (dict, optional) – A dictionary of parameters and values to be added to the results class. Default is None.
Returns: results – Returns a list of Results objects one per each train/test fraction and each node shuffle.
Return type: list
-
class
evalne.evaluation.evaluator.NREvaluator(traintest_split, dim=128, lp_model=LogisticRegressionCV(cv=5, scoring='roc_auc'))[source]¶ Bases:
evalne.evaluation.evaluator.LPEvaluatorClass designed to simplify the evaluation of embedding methods for network reconstruction tasks. The train graph is assumed to be the entire network. Parameter tuning is performed directly on this complete graph.
Parameters: - traintest_split (NREvalSplit) – An object containing the train graph (in this case the full network) and a set of train edges and non-edges. These edges can be all edges in the graph or a subset.
- dim (int, optional) – Embedding dimensionality. Default is 128.
- lp_model (Sklearn binary classifier, optional) – The binary classifier to use for prediction. Default is logistic regression with 5 fold cross validation: LogisticRegressionCV(Cs=10, cv=5, penalty=’l2’, scoring=’roc_auc’, solver=’lbfgs’, max_iter=100))
Notes
In network reconstruction the aim is to asses how well an embedding method captures the structure of a given graph. The embedding methods are trained on a complete input graph. Hyperparameter tuning is performed directly on this graph (overfitting is, in this case, expected and desired). The embeddings obtained are used to perform link predictions and their quality is evaluated. Checking the link predictions for all node pairs is generally unfeasible, therefore a subset of all node pairs in the input graph are selected for evaluation.
Examples
Instantiating an NREvaluator with default parameters (for this task train/validation splits are not necessary):
>>> from evalne.evaluation.evaluator import NREvaluator >>> from evalne.evaluation.split import NREvalSplit >>> from evalne.utils import preprocess as pp >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Create the required train/test split >>> traintest_split = NREvalSplit() >>> _ = traintest_split.compute_splits(G) >>> # Initialize the NREvaluator >>> nee = NREvaluator(traintest_split)
Instantiating an NREvaluator where we randomly select 10% of all node pairs in the network for evaluation:
>>> from evalne.evaluation.evaluator import NREvaluator >>> from evalne.evaluation.split import NREvalSplit >>> from evalne.utils import preprocess as pp >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Create the required train/test split and sample 0.1, i.e. 10% of all nodes >>> traintest_split = NREvalSplit() >>> _ = traintest_split.compute_splits(G, samp_frac=0.1) >>> # Initialize the NREvaluator >>> nee = NREvaluator(traintest_split)
-
evaluate_cmd(method_name, method_type, command, edge_embedding_methods, input_delim, output_delim, tune_params=None, maximize='auroc', write_weights=False, write_dir=False, timeout=None, verbose=True)[source]¶ Evaluates an embedding method and tunes its parameters from the method’s command line call string. This function can evaluate node embedding, node-pair embedding or end to end predictors. If model parameter tuning is required, models are tuned directly on the train data. The returned Results object will only contain train scores.
Parameters: - method_name (string) – A string indicating the name of the method to be evaluated.
- method_type (string) – A string indicating the type of embedding method (i.e. ne, ee, e2e). NE methods are expected to return embeddings, one per graph node, as either dict or matrix sorted by nodeID. EE methods are expected to return node-pair emb. as [num_edges x embed_dim] matrix in same order as input. E2E methods are expected to return predictions as a vector in the same order as the input edgelist.
- command (string) – A string containing the call to the method as it would be written in the command line. For ‘ne’ methods placeholders (i.e. {}) need to be provided for the parameters: input network file, output file and embedding dimensionality, precisely IN THIS ORDER. For ‘ee’ methods with parameters: input network file, input train edgelist, input test edgelist, output train embeddings, output test embeddings and embedding dimensionality, 6 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER. For methods with parameters: input network file, input edgelist, output embeddings, and embedding dimensionality, 4 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER. For ‘e2e’ methods with parameters: input network file, input train edgelist, input test edgelist, output train predictions, output test predictions and embedding dimensionality, 6 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER. For methods with parameters: input network file, input edgelist, output predictions, and embedding dimensionality, 4 placeholders (i.e. {}) need to be provided, precisely IN THIS ORDER.
- edge_embedding_methods (array-like) – A list of methods used to compute node-pair embeddings from the node embeddings output by NE models. The accepted values are the function names in evalne.evaluation.edge_embeddings. When evaluating ‘ee’ or ‘e2e’ methods, this parameter is ignored.
- input_delim (string) – The delimiter expected by the method as input (edgelist).
- output_delim (string) – The delimiter provided by the method in the output.
- tune_params (string, optional) – A string containing all the parameters to be tuned and their values. Default is None.
- maximize (string, optional) – The score to maximize while performing parameter tuning. Default is ‘auroc’.
- write_weights (bool, optional) – If True the train graph passed to the embedding methods will be stored as weighted edgelist (e.g. triplets src, dst, weight) otherwise as normal edgelist. If the graph edges have no weight attribute and this parameter is set to True, a weight of 1 will be assigned to each edge. Default is False.
- write_dir (bool, optional) – This option is only relevant for undirected graphs. If False, the train graph will be stored with a single direction of the edges. If True, both directions of edges will be stored. Default is False.
- timeout (float or None, optional) – A float indicating the maximum amount of time (in seconds) the evaluation can run for. If None, the evaluation is allowed to continue until completion. Default is None.
- verbose (bool, optional) – A parameter to control the amount of screen output. Default is True.
Returns: results – The evaluation results as a Results object.
Return type: Raises: TimeoutExpired– If the execution does not finish within the allocated time.IOError– If the method call does not succeed.ValueError– If the method type is unknown. If for a method all parameter combinations fail to provide results.
See also
evalne.utils.util.run()- The low level function used to run a cmd call with given timeout.
Examples
Evaluating the OpenNE implementation of node2vec without parameter tuning and with ‘average’ and ‘hadamard’ as node-pair embedding operators. We assume the method is installed in a virtual environment and that an evaluator (nee) has already been instantiated (see class examples):
>>> # Prepare the cmd command for running the method. If running on a python console full paths are required >>> cmd = '../OpenNE-master/venv/bin/python -m openne --method node2vec ' ... '--graph-format edgelist --input {} --output {} --representation-size {}' >>> # Call the evaluation >>> result = nee.evaluate_cmd(method_name='Node2vec', method_type='ne', command=cmd, ... edge_embedding_methods=['average', 'hadamard'], input_delim=' ', output_delim=' ') Running command... [...] >>> # Print the results >>> result.pretty_print() Method: Node2vec Parameters: [('split_id', 0), ('dim', 128), ('eval_time', 21.773473024368286), ('nw_name', 'test'), ('split_alg', 'random_edge_sample'), ('train_frac', 1), ('edge_embed_method', 'hadamard'), ('samp_frac', 0.01)] Train scores: tn = 2444 [...]
Evaluating the metapath2vec c++ implementation with parameter tuning and with ‘average’ node-pair embedding operator. We assume the method is installed and that an evaluator (nee) has already been instantiated (see class examples):
>>> # Prepare the cmd command for running the method. If running on a python console full paths are required >>> cmd = '../../methods/metapath2vec/metapath2vec -min-count 1 -iter 20 ' ... '-samples 100 -train {} -output {} -size {}' >>> # Call the evaluation >>> result = nee.evaluate_cmd(method_name='Metapath2vec', method_type='ne', command=cmd, ... edge_embedding_methods=['average'], input_delim=' ', output_delim=' ') Running command... [...] >>> # Print the results >>> result.pretty_print() Method: Metapath2vec Parameters: Method: Metapath2vec Parameters: [('split_id', 0), ('dim', 128), ('eval_time', 1.948814868927002), ('nw_name', 'test'), ('split_alg', 'random_edge_sample'), ('train_frac', 1), ('edge_embed_method', 'average'), ('samp_frac', 0.01)] Train scores: tn = 2444 [...]
-
class
evalne.evaluation.evaluator.SPEvaluator(traintest_split, trainvalid_split=None, dim=128, lp_model=LogisticRegressionCV(cv=5, scoring='roc_auc'))[source]¶ Bases:
evalne.evaluation.evaluator.LPEvaluatorClass designed to simplify the evaluation of embedding methods for sign prediction tasks. The train and validation graphs are assumed to be weighted and contain positive and negative edges. This is a simple extension of an LP evaluation which overrides the baseline implementation to work for sign prediction.
Parameters: - traintest_split (SPEvalSplit) – An object containing the train graph (a subgraph of the full network that spans all nodes) and a set of train positive and negative edges. Test edges are optional. If not provided only train results will be generated.
- trainvalid_split (SPEvalSplit, optional) – An object containing the validation graph (a subgraph of the training network that spans all nodes) and a set of positive and negative edges. If not provided a split with the same parameters as the train one, but with train_frac=0.9, will be computed. Default is None.
- dim (int, optional) – Embedding dimensionality. Default is 128.
- lp_model (Sklearn binary classifier, optional) – The binary classifier to use for prediction. Default is logistic regression with 5 fold cross validation: LogisticRegressionCV(Cs=10, cv=5, penalty=’l2’, scoring=’roc_auc’, solver=’lbfgs’, max_iter=100)).
Notes
In sign prediction the aim is to predict the sign (positive or negative) of given edges. The existence of the edges is assumed (i.e. we do not predict the sign of unconnected node pairs). Therefore, sign prediction is also a binary classification task similar to link prediction where, instead of predicting the existence of edges or not, we predict the signs for edges we know exist. Unlike for link prediction, in this case we do not need to perform negative sampling, since we already have both classes (the positively and the negatively connected node pairs).
Examples
Instantiating an SPEvaluator without a specific train/validation split (this split will be computed automatically if parameter tuning for any method is required):
>>> from evalne.evaluation.evaluator import SPEvaluator >>> from evalne.evaluation.split import SPEvalSplit >>> from evalne.utils import preprocess as pp >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/sig_network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Create the required train/test split >>> traintest_split = SPEvalSplit() >>> _ = traintest_split.compute_splits(G) >>> # Check that the train test parameters are indeed the correct ones >>> traintest_split.get_parameters() {'split_id': 0, 'nw_name': 'test', 'split_alg': 'spanning_tree', 'train_frac': 0.4980} >>> # Initialize the SPEvaluator >>> nee = SPEvaluator(traintest_split)
Instantiating an SPEvaluator with a specific train/validation split (allows the user to specify any parameters for the train/validation split). Use ‘fast’ as the algorithm to split train and test edges and set train fraction to 0.8 for both train and validation splits:
>>> from evalne.evaluation.evaluator import SPEvaluator >>> from evalne.evaluation.split import SPEvalSplit >>> from evalne.utils import preprocess as pp >>> # Load and preprocess a network >>> G = pp.load_graph('./evalne/tests/data/sig_network.edgelist') >>> G, _ = pp.prep_graph(G) >>> # Create the required train/test split >>> traintest_split = SPEvalSplit() >>> _ = traintest_split.compute_splits(G, train_frac=0.8, split_alg='fast') >>> # Check that the train test parameters are indeed the correct ones >>> traintest_split.get_parameters() {'split_id': 0, 'nw_name': 'test', 'split_alg': 'fast', 'train_frac': 0.8125} >>> # Create the train/validation split from the train data computed in the trintest_split >>> # The graph used to initialize this split must, thus, be the train graph from the traintest_split >>> trainvalid_split = SPEvalSplit() >>> _ = trainvalid_split.compute_splits(traintest_split.TG, train_frac=0.8, split_alg='fast') >>> # Initialize the SPEvaluator >>> nee = SPEvaluator(traintest_split, trainvalid_split)
-
evaluate_baseline(method, neighbourhood='in', timeout=None)[source]¶ Evaluates the baseline method requested. Evaluation output is returned as a Results object. To evaluate the baselines on sign prediction we remove all negative edges from the train graph in traintest_split. For Katz neighbourhood=`in` and neighbourhood=`out` will return the same results corresponding to neighbourhood=`in`. Execution time is contained in the results object. If the train/test split object used to initialize the evaluator does not contain test edges, the results object will only contain train results.
Parameters: - method (string) – A string indicating the name of any baseline from evalne.methods to evaluate.
- neighbourhood (string, optional) – A string indicating the ‘in’ or ‘out’ neighbourhood to be used for directed graphs. Default is ‘in’.
- timeout (float or None) – A float indicating the maximum amount of time (in seconds) the evaluation can run for. If None, the evaluation is allowed to continue until completion. Default is None.
Returns: results – The evaluation results as a Results object.
Return type: Raises: TimeoutExpired– If the execution does not finish within the allocated time.TypeError– If the Katz method call is incorrect.ValueError– If the heuristic selected does not exist.
See also
evalne.utils.util.run_function()- The low level function used to run a baseline with given timeout.
Examples
Evaluating the common neighbours heuristic with default parameters. We assume an evaluator (nee) has already been instantiated (see class examples):
>>> result = nee.evaluate_baseline(method='common_neighbours') >>> # Print the results >>> result.pretty_print() Method: common_neighbours Parameters: [('split_id', 0), ('dim', 128), ('neighbourhood', 'in'), ('split_alg', 'fast'), ('eval_time', 0.04459214210510254), ('nw_name', 'test'), ('train_frac', 0.8125)] Test scores: tn = 71 [...]
Evaluating katz with beta=0.05 and timeout 60 seconds. We assume an evaluator (nee) has already been instantiated (see class examples):
>>> result = nee.evaluate_baseline(method='katz 0.05', timeout=60) >>> # Print the results >>> result.pretty_print() Method: katz 0.05 Parameters: [('split_id', 0), ('dim', 128), ('neighbourhood', 'in'), ('split_alg', 'fast'), ('eval_time', 0.1246330738067627), ('nw_name', 'test'), ('train_frac', 0.8125)] Test scores: tn = 120 [...]
evalne.evaluation.pipeline module¶
-
class
evalne.evaluation.pipeline.EvalSetup(configpath, run_checks=True)[source]¶ Bases:
objectClass that acts as a wrapper for the EvalNE .ini configuration files. Options (or variables) in the .ini files are exposed as class properties and basic input checks are performed.
Parameters: - configpath (string) – The path of the .ini configuration file.
- run_checks (bool, optional) – Toggles .ini file parameter checks. Default is True.
-
comments¶ Returns a list of strings, the characters denoting comments in the network files.
-
curves¶ Returns a string indicating the curves to provide as output.
-
del_selfloops¶ Returns a bool, delete or not self loops in the network.
-
delimiter¶ Returns a string indicating the delimiter to be used when writing the preprocessed graphs to a files.
-
directed¶ Returns a bool indicating if all the networks are directed or not.
-
edge_embedding_methods¶ {‘average’, ‘hadamard’, ‘weighted_l1’, ‘weighted_l2’}
Type: Returns a list of strings indicating the node-pair operators to use. Possible values
-
embed_dim¶ Returns an int indicating the dimensions of the embedding.
-
embtype_other¶ node embeddings (ne), edge embeddings (ee) or node similarities (e2e). Possible values: {‘ne’, ‘ee’, ‘e2e’}.
Type: Returns a list of strings indicating the method’s output type
-
fe_ratio¶ Returns a float indicating the ratio of non-edges to edges for tr & te. The num_fe = fe_ratio * num_edges.
-
getboollist(section, option)[source]¶ Reads a string option and returns it as a list of booleans. The input string is split by any kind of white space separator. Elements such as ‘True’, ‘true’, ‘1’, ‘yes’, ‘on’ are mapped to True. Elements such as ‘False’, ‘false’, ‘0’, ‘no’, ‘off’ are mapped to False.
Parameters: - section (string) – A config file section name.
- option (string) – A config file option name.
Returns: list – A list of booleans.
Return type: list
-
getlinelist(section, option)[source]¶ Reads a string option and returns it as a list of strings split by new lines only.
Parameters: - section (string) – A config file section name.
- option (string) – A config file option name.
Returns: list – A list of strings.
Return type: list
-
getlist(section, option, dtype)[source]¶ Reads a string option and returns it as a list of elements of the specified type. The input string is split by any kind of white space separator.
Parameters: - section (string) – A config file section name.
- option (string) – A config file option name.
- dtype (primitive type) – The desired type of the elements in the output list.
Returns: list – A list of elements cast to the specified primitive type.
Return type: list
-
getseplist(section, option)[source]¶ Reads a string option containing several separators (‘s’, ‘t’ and ‘n’ ) and returns it as a list of proper string separators (white space, tab or new line).
Parameters: - section (string) – A config file section name.
- option (string) – A config file option name.
Returns: list – A list of strings.
Return type: list
-
gettuneparams(library)[source]¶ Reads a ‘TUNE_PARAMS’ option that contain parameters and their associated values (e.g. ‘TUNE_PARAMS’). The method returns the option as a list of strings split by new lines. The list if filled with None if needed so the length is the same as the number of methods being evaluated.
Parameters: library (string) – A string indicating if the openne or other ‘TUNE_PARAMS’ should be checked. Accepted values are: ‘opne’, ‘other’. Returns: tune_params – A list of string or None containing parameters and their values. Return type: list
-
inpaths¶ Returns a list of strings indicating the paths to files containing the networks. A check is performed to ensure the paths exist.
-
input_delim_other¶ Returns a list of strings indicating the input delimiters expected the by each methods.
-
labelpaths¶ Returns a list of string indicating the paths where the node label files can be found. Required if task is ‘nc’
-
lp_baselines¶ {‘’, ‘random_prediction’, ‘common_neighbours’, ‘jaccard_coefficient’, ‘adamic_adar_index’, ‘preferential_attachment’, ‘resource_allocation_index’, ‘cosine_similarity’, ‘lhn_index’, ‘topological_overlap’, ‘katz’, ‘all_baselines’}
Type: Returns a list of strings indicating the link prediction heuristics to evaluate. Possible values
-
lp_model¶ Returns an sklearn binary classifier used to predict links from node-pair embeddings.
-
lp_num_edge_splits¶ Returns an int indicating the number of repetitions for experiment with different train/test edge splits. Required if task is ‘lp’ or ‘sp’. For ‘nr’ and ‘nc’ this value must be 1.
-
maximize¶ Returns a string indicating the score to maximize when performing model validation. Possible values for LP, SP and NR: {‘auroc’, ‘f_score’, ‘precision’, ‘recall’, ‘accuracy’, ‘fallout’, ‘miss’}. Possible values for NC: {‘f1_micro’, ‘f1_macro’, ‘f1_weighted’}
-
methods_opne¶ Returns a list of strings indicating the command line calls to perform in order to evaluate each method.
-
methods_other¶ Returns a list of strings indicating the command line calls to perform in order to evaluate each method.
-
names¶ Returns a list of strings indicating the names of the networks to be evaluated.
-
names_opne¶ Returns a list of strings indicating the names of methods from OpenNE to be evaluated. In the same order as METHODS_OPNE.
-
names_other¶ Returns a list of strings indicating the names of any other methods not from OpenNE to be evaluated. In the same order as METHODS_OTHER.
-
nc_node_fracs¶ Returns a list of float indicating the fractions of train labels to use when evaluating NC. Required if task is ‘nc’.
-
nc_num_node_splits¶ Returns an int indicating the number of repetitions for NC experiments with different train/test node splits. Required if task is ‘nc’.
-
neighbourhood¶ Returns a list of string indicating, for directed graphs, if the in or the out neighbourhood should be used. Possible values: {‘’, ‘in’, ‘out’}
-
nr_edge_samp_frac¶ Returns a float indicating the fraction of all possible node pairs to sample and compute precision@k for when evaluating NR. Required if task is ‘nr’.
-
output_delim_other¶ Returns a list of strings indicating the delimiter used by each method in the output file (when writing node embeddings, edge embeddings or predictions).
-
output_format_other¶ Returns
-
owa¶ Returns a bool, indicating if the open world (True) or the closed world assumption (False) for non-edges should be used.
-
precatk_vals¶ Returns a list of int indicating the values of k for which to provide the precision at k.
-
relabel¶ Returns a bool, relabel or not the network nodes to 0…N (required for methods such as PRUNE)
-
save_prep_nw¶ Returns a bool if the preprocessed graph should be stored or not.
-
scores¶ {‘’, ‘%(maximize)s’, ‘all’}
Type: Returns a string indicating the score to be reported in the output file. Possible values
-
seed¶ {‘’, ‘None’, any_int}
Type: Returns and int or None indicating the random seed to use in the experiments. Possible values
-
separators¶ Returns a list of strings indicating the separators used in the network files.
-
split_alg¶ Returns a string indicating the algorithm to use for splitting edges in train/test, train/validation sets. Possible values: {‘spanning_tree’, ‘random’, ‘naive’, ‘fast’, ‘timestamp’}.
-
task¶ Returns a string indicating the task to evaluate i.e. link prediction (LP), sign prediction (SP), network reconstruction (NR) or node classification (NC). Possible values: {‘lp’, ‘sp’, ‘nr’, ‘nc’}
-
timeout¶ Returns a float indicating the maximum execution time in seconds (or None) for each method including hyperparameter tuning.
-
traintest_frac¶ Returns a float indicating the fraction of total edges to use for training and validation. The rest should be used for testing.
-
trainvalid_frac¶ Returns a float indicating the fraction of train-validation edges to use for training. The rest should be used for validation.
-
tune_params_opne¶ Returns a list of strings indicating the parameters of methods from OpenNE to be tuned by the library and values to try.
-
tune_params_other¶ Returns a list of strings indicating the parameters to be tuned by the library.
-
verbose¶ Returns a bool indicating the verbosity level of the execution.
-
write_dir_other¶ Returns a list of bool indicating if training graphs should be given as input to methods with both edge dir. (True) or one (False).
-
write_stats¶ Returns a bool, write or not common graph statistics as header in the preprocessed network file.
-
write_weights_other¶ Returns a list of bool indicating if training graphs should be given as input to methods weighted (True) or unweighted (False).
evalne.evaluation.score module¶
-
class
evalne.evaluation.score.NCResults(method, params, train_pred, train_labels, test_pred=None, test_labels=None)[source]¶ Bases:
objectClass that encapsulates the train and test predictions of one method on a specific network and set of parameters. The train and test predictions are stored as NCScores objects. Functions for plotting, printing and saving to files the train and test scores are provided. Supports multi-label classification.
Parameters: - method (string) – A string representing the name of the method associated with these results.
- params (dict) – A dictionary of parameters used to obtain these results. Includes wall clock time of method evaluation.
- train_pred (ndarray) – An array containing the train predictions.
- train_labels (ndarray) – An array containing the train labels.
- test_pred (ndarray, optional) – An array containing the test predictions. Default is None.
- test_labels (ndarray, optional) – An array containing the test labels. Default is None.
Variables: - method (string) – A string representing the name of the method associated with these results.
- params (dict) – A dictionary of parameters used to obtain these results. Includes wall clock time of method evaluation.
- train_scores (Scores) – An NCScores object containing train scores.
- test_scores (Scores, optional) – An NCScores object containing test scores. Default is None.
-
get_all(results='auto', precatk_vals=None)[source]¶ Returns the names of all performance metrics that can be computed from train or test predictions and their associated values. These metrics are: ‘f1_micro’, ‘f1_macro’, ‘f1_weighted’.
Parameters: - results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
- precatk_vals (None, optional) – Not used.
Raises: ValueError– If test results are requested but not initialized in constructor.
-
pretty_print(results='auto')[source]¶ Prints to screen the method name, execution parameters, and all available performance metrics (for train or test predictions).
Parameters: results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise). Raises: ValueError– If test results are requested but not initialized in constructor.See also
get_all()- Describes all the performance metrics that can be computed from train or test predictions.
-
save(filename, results='auto')[source]¶ Writes the method name, execution parameters, and all available performance metrics (for train or test predictions) to a file.
Parameters: - filename (string or file) – A file or filename where to store the output.
- results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
Raises: ValueError– If test results are required but not initialized in constructor.See also
get_all()- Describes all the performance metrics that can be computed from train or test predictions.
-
save_predictions(filename, results='auto')[source]¶ Writes the method name, execution parameters, and the train or test predictions to a file.
Parameters: - filename (string or file) – A file or filename where to store the output.
- results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
Raises: ValueError– If test results are required but not initialized in constructor.
-
class
evalne.evaluation.score.NCScores(y_true, y_pred)[source]¶ Bases:
objectClass that encapsulates train or test predictions and exposes methods to compute different performance metrics. Supports multi-label classification.
Parameters: - y_true (ndarray) – An array containing the true labels.
- y_pred (ndarray) – An array containing the predictions.
Variables: - y_true (ndarray) – An array containing the true labels.
- y_pred (ndarray) – An array containing the predictions.
-
f1_macro()[source]¶ Computes the f1 score for each label, and finds their unweighted average. This metric does not take label imbalance into account.
Returns: f1_macro – The f1 macro score. Return type: float
-
class
evalne.evaluation.score.Results(method, params, train_pred, train_labels, test_pred=None, test_labels=None, label_binarizer=LogisticRegression(solver='liblinear'))[source]¶ Bases:
objectClass that encapsulates the train and test predictions of one method on a specific network and set of parameters. The train and test predictions are stored as Scores objects. Functions for plotting, printing and saving to files the train and test scores are provided. Supports binary classification only.
Parameters: - method (string) – A string representing the name of the method associated with these results.
- params (dict) – A dictionary of parameters used to obtain these results. Includes wall clock time of method evaluation.
- train_pred (ndarray) – An array containing the train predictions.
- train_labels (ndarray) – An array containing the train labels.
- test_pred (ndarray, optional) – An array containing the test predictions. Default is None.
- test_labels (ndarray, optional) – An array containing the test labels. Default is None.
- label_binarizer (string or Sklearn binary classifier, optional) – If the predictions returned by the model are not binary, this parameter indicates how these binary predictions should be computed in order to be able to provide metrics such as the confusion matrix. Any Sklear binary classifier can be used or the keyword ‘median’ which will used the prediction medians as binarization thresholds. Default is LogisticRegression(solver=’liblinear’)
Variables: - method (string) – A string representing the name of the method associated with these results.
- params (dict) – A dictionary of parameters used to obtain these results. Includes wall clock time of method evaluation.
- binary_preds (bool) – A bool indicating if the train and test predictions are binary or not.
- train_scores (Scores) – A Scores object containing train scores.
- test_scores (Scores, optional) – A Scores object containing test scores. Default is None.
- label_binarizer (string or Sklearn binary classifier, optional) – If the predictions returned by the model are not binary, this parameter indicates how these binary predictions should be computed in order to be able to provide metrics such as the confusion matrix. By default, the method binarizes the predictions such that their accuracy is maximised. Any Sklearn binary classifier can be used or the keyword ‘median’ which will used the prediction medians as binarization thresholds. Default is LogisticRegression(solver=’liblinear’)
Raises: AttributeError– If the label binarizer is set to an incorrect value.-
get_all(results='auto', precatk_vals=None)[source]¶ Returns the names of all performance metrics that can be computed from train or test predictions and their associated values. These metrics are: ‘tn’, ‘fp’, ‘fn’, ‘tp’, ‘auroc’, ‘average_precision’, ‘precision’, ‘precisionatk’, ‘recall’, ‘fallout’, ‘miss’, ‘accuracy’ and ‘f_score’.
Parameters: - results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
- precatk_vals (list of int or None, optional) – The values for which the precision at k should be computed. Default is None.
Raises: ValueError– If test results are requested but not initialized in constructor.
-
plot(filename=None, results='auto', curve='all')[source]¶ Plots PR or ROC curves of the train or test predictions. If a filename is provided, the method will store the plot in pdf format to a file named <filename>+’_PR.pdf’ or <filename>+’_ROC.pdf’.
Parameters: - filename (string, optional) – A string indicating the path and name of the file where to store the plot. If None, the plots are only shown on screen. Default is None.
- results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
- curve (string, optional) – Can be one of ‘all’, ‘pr’ or ‘roc’. Default is ‘all’ (generates both curves).
Raises: ValueError– If test results are requested but not initialized in constructor.
-
pretty_print(results='auto', precatk_vals=None)[source]¶ Prints to screen the method name, execution parameters, and all available performance metrics (for train or test predictions).
Parameters: - results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
- precatk_vals (list of int or None, optional) – The values for which the precision at k should be computed. Default is None.
Raises: ValueError– If test results are requested but not initialized in constructor.See also
get_all()- Describes all the performance metrics that can be computed from train or test predictions.
-
save(filename, results='auto', precatk_vals=None)[source]¶ Writes the method name, execution parameters, and all available performance metrics (for train or test predictions) to a file.
Parameters: - filename (string or file) – A file or filename where to store the output.
- results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
- precatk_vals (list of int or None, optional) – The values for which the precision at k should be computed. Default is None.
Raises: ValueError– If test results are required but not initialized in constructor.See also
get_all()- Describes all the performance metrics that can be computed from train or test predictions.
-
save_predictions(filename, results='auto')[source]¶ Writes the method name, execution parameters, and the train or test predictions and corresponding labels to a file.
Parameters: - filename (string or file) – A file or filename where to store the output.
- results (string, optional) – A string indicating if the ‘train’ or ‘test’ predictions should be used. Default is ‘auto’ (selects ‘test’ if test predictions are logged and ‘train’ otherwise).
Raises: ValueError– If test results are required but not initialized in constructor.
-
class
evalne.evaluation.score.Scores(y_true, y_pred, y_bin)[source]¶ Bases:
objectClass that encapsulates train or test predictions and exposes methods to compute different performance metrics. Supports binary classification only.
Parameters: - y_true (ndarray) – An array containing the true labels.
- y_pred (ndarray) – An array containing the predictions.
- y_bin (ndarray) – An array containing binarized predictions.
Variables: - y_true (ndarray) – An array containing the true labels.
- y_pred (ndarray) – An array containing the predictions.
- y_bin (ndarray) – An array containing binarized predictions.
- tn (float) – The number of true negative in prediction.
- fp (float) – The number of false positives in prediction.
- fn (float) – The number of false negatives in prediction.
- tp (float) – The number of true positives in prediction.
-
accuracy()[source]¶ Computes the accuracy score.
Returns: accuracy – The prediction accuracy score. Return type: float
-
auroc()[source]¶ Computes the Area Under the Receiver Operating Characteristic Curve (ROC AUC).
Returns: auroc – The prediction auroc score. Return type: float Notes
Throws a warning if class imbalance is detected.
-
average_precision()[source]¶ Computes the average precision score.
Returns: avgprec – The average precision score. Return type: float
-
f_score(beta=1)[source]¶ Computes the F-score as the weighted harmonic mean of precision and recall.
Parameters: beta (float, optional) – Allows to assign more weight to precision or recall. If beta > 1, recall is emphasized over precision. If beta < 1, precision is emphasized over recall. Returns: f_score – The prediction f_score. Return type: float Notes
The generalized form is used, where P and R represent precision and recall, respectively:
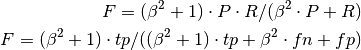
-
fallout()[source]¶ Computes the fallout in prediction.
Returns: fallout – The prediction fallout score. Return type: float
-
miss()[source]¶ Computes the miss in prediction.
Returns: miss – The prediction miss score. Return type: float
-
precision()[source]¶ Computes the precision in prediction.
Returns: precision – The prediction precision score. Return type: float
-
class
evalne.evaluation.score.Scoresheet(tr_te='test', precatk_vals=None)[source]¶ Bases:
objectClass that simplifies the logging and management of evaluation results and execution times. Functions for logging, plotting and writing the results to files are provided. The Scoresheet does not log the complete train or test model predictions.
Parameters: - tr_te (string, optional) – A string indicating if the ‘train’ or ‘test’ results should be stored. Default is ‘test’.
- precatk_vals (list of int or None, optional) – The values for which the precision at k should be computed. Default is None.
-
get_latex(metric='auroc')[source]¶ Returns a view of the Scoresheet as a Latex table for the specified metric. The columns of the table represent different networks and the rows different methods. If multiple Results for the same network/method combination were logged (multiple repetitions of the experiment), the average is returned.
Parameters: metric (string, optional) – Can be one of ‘tn’, ‘fp’, ‘fn’, ‘tp’, ‘auroc’, ‘average_precision’, ‘precision’, ‘recall’, ‘fallout’, ‘miss’, ‘accuracy’, ‘f_score’, ‘eval_time’ or ‘edge_embed_method’. Default is ‘auroc’. Returns: latex_table – A latex table as a string. Return type: string
-
get_pandas_df(metric='auroc', repeat=None)[source]¶ Returns a view of the Scoresheet as a pandas DataFrame for the specified metric. The columns of the DataFrame represent different networks and the rows different methods. If multiple Results for the same network/method combination were logged (multiple repetitions of the experiment), one can select any of these repeats or get the average over all.
Parameters: - metric (string, optional) – Can be one of ‘tn’, ‘fp’, ‘fn’, ‘tp’, ‘auroc’, ‘average_precision’, ‘precision’, ‘recall’, ‘fallout’, ‘miss’, ‘accuracy’, ‘f_score’, ‘eval_time’ or ‘edge_embed_method’. Default is ‘auroc’.
- repeat (int, optional) – An int indicating the experiment repeat for which the results should be returned. If not indicated, the average over all repeats will be computed and returned. Default is None (computes average over repeats).
Returns: df – A pandas DataFrame view of the Scoresheet for the specified metric.
Return type: DataFrame
Raises: ValueError– If the requested metric does not exist. If the Scoresheet is empty so a DataFrame can not be generated.Notes
For uncountable ‘metrics’ such as the node pair embedding operator (i.e ‘edge_embed_method’), avg returns the most frequent item in the vector.
Examples
Read a scoresheet and get the auroc scores as a pandas DataFrame
>>> scores = pickle.load(open('lp_eval_1207_1638/eval.pkl', 'rb')) >>> df = scores.get_pandas_df() >>> df Network_1 Network_2 katz 0.8203 0.8288 common_neighbours 0.3787 0.3841 jaccard_coefficient 0.3787 0.3841
Read a scoresheet and get the f scores of the first repetition of the experiment
>>> scores = pickle.load(open('lp_eval_1207_1638/eval.pkl', 'rb')) >>> df = scores.get_pandas_df('f_score', repeat=0) >>> df Network_1 Network_2 katz 0 0 common_neighbours 0.7272 0.7276 jaccard_coefficient 0.7265 0.7268
-
log_results(results)[source]¶ Logs in the Scoresheet all the performance metrics (and execution time) extracted from the input Results object or list of Results objects. Multiple Results for the same method on the same network can be provided and will all be stored (these are assumed to correspond to different repetitions of the experiment).
Parameters: results (Results or list of Results) – The Results object or objects to be logged in the Scoresheet. Examples
Evaluate the common neighbours baseline and log the train and test results:
>>> tr_scores = Scoresheet(tr_te='train') >>> te_scores = Scoresheet(tr_te='test') >>> result = nee.evaluate_baseline(method='common_neighbours') >>> tr_scores.log_results(result) >>> te_scores.log_results(result)
-
print_tabular(metric='auroc')[source]¶ Prints a tabular view of the Scoresheet for the specified metric. The columns of the table represent different networks and the rows different methods. If multiple Results for the same network/method combination were logged (multiple repetitions of the experiment), the average is showed.
Parameters: metric (string, optional) – Can be one of ‘tn’, ‘fp’, ‘fn’, ‘tp’, ‘auroc’, ‘average_precision’, ‘precision’, ‘recall’, ‘fallout’, ‘miss’, ‘accuracy’, ‘f_score’, ‘eval_time’ or ‘edge_embed_method’. Default is ‘auroc’. Examples
Read a scoresheet and get the average execution times over all experiment repeats as tabular output:
>>> scores = pickle.load(open('lp_eval_1207_1638/eval.pkl', 'rb')) >>> scores.print_tabular('eval_time') Network_1 Network_2 katz 0.0350 0.0355 common_neighbours 0.0674 0.0676 jaccard_coefficient 0.6185 0.6693
-
write_all(filename, repeats='avg')[source]¶ Writes for all networks, methods and performance metrics the corresponding values to a file. If multiple Results for the same network/method combination were logged (multiple repetitions of the experiment), the method can return the average or all logged values.
Parameters: - filename (string) – A file where to store the results.
- repeats (string, optional) – Can be one of ‘all’, ‘avg’. Default is ‘avg’.
Notes
For uncountable ‘metrics’ such as the node pair embedding operator (i.e ‘edge_embed_method’), avg returns the most frequent item in the vector.
Examples
Read a scoresheet and write all metrics to a file with repeats=’avg’:
>>> scores = pickle.load(open('lp_eval_1207_1638/eval.pkl', 'rb')) >>> scores.write_all('./test.txt') >>> print(open('test.txt', 'rb').read()) Network_1 Network --------------------------- katz: tn: 684.0 fp: 0.0 fn: 684.0 tp: 0.0 auroc: 0.8203 ...
Read a scoresheet and write all metrics to a file with repeats=’all’:
>>> scores = pickle.load(open('lp_eval_1207_1638/eval.pkl', 'rb')) >>> scores.write_all('./test.txt', 'all') >>> print(open('test.txt', 'rb').read()) Network_1 Network --------------------------- katz: tn: [684 684] fp: [0 0] fn: [684 684] tp: [0 0] auroc: [0.8155 0.8252] ...
-
write_pickle(filename)[source]¶ Writes a pickle representation of this object to a file.
Parameters: filename (string) – A file where to store the pickle representation.
-
write_tabular(filename, metric='auroc')[source]¶ Writes a tabular view of the Scoresheet for the specified metric to a file. The columns of the table represent different networks and the rows different methods. If multiple Results for the same network/method combination were logged (multiple repetitions of the experiment), the average is used.
Parameters: - filename (string) – A file where to store the results.
- metric (string, optional) – Can be one of ‘tn’, ‘fp’, ‘fn’, ‘tp’, ‘auroc’, ‘average_precision’, ‘precision’, ‘recall’, ‘fallout’, ‘miss’, ‘accuracy’, ‘f_score’ or ‘eval_time’. Default is ‘auroc’.
evalne.evaluation.split module¶
-
class
evalne.evaluation.split.BaseEvalSplit[source]¶ Bases:
objectBase class that provides a high level interface for managing/computing sets of train and test edges and non-edges for LP, SP and NR tasks. The class exposes the train edges and non-edges through the train_edges property and the test edges and non-edges through the test_edges property. Parameters used to compute these sets are also made available.
-
TG¶ A NetworkX graph or digraph to be used for training the embedding methods. For LP this should be the graph spanned by all train edges, for SP the graph spanned by the positive and negative train edges (with signs as edge weights) and for NR the entire graph being evaluated.
-
get_data()[source]¶ Returns the sets of train and test node pairs and label vectors.
Returns: - train_edges (set) – Set of all train edges and non-edges.
- test_edges (set) – Set of all test edges and non-edges.
- train_labels (list) – A list of labels indicating if each train node-pair is an edge or non-edge (1 or 0).
- test_labels (list) – A list of labels indicating if each test node-pair is an edge or non-edge (1 or 0).
-
get_parameters()[source]¶ Returns the class properties except the sets of train and test node pairs, labels and train graph.
Returns: parameters – The parameters used when computing this split as a dictionary of parameters and values. Return type: dict
-
nw_name¶ A string indicating the name of the dataset used to generate the sets of edges.
-
save_tr_graph(output_path, delimiter, write_stats=False, write_weights=False, write_dir=True)[source]¶ Saves the TG graph to a file.
Parameters: - output_path (file or string) – File or filename to write. If a file is provided, it must be opened in ‘wb’ mode.
- delimiter (string, optional) – The string used to separate values. Default is ‘,’.
- write_stats (bool, optional) – Adds basic graph statistics to the file as a header or not. Default is True.
- write_weights (bool, optional) – If True data will be stored as weighted edgelist i.e. triplets (src, dst, weight), otherwise, as regular (src, dst) pairs. For unweighted graphs, setting this parameter to True will add weight 1 to all edges. Default is False.
- write_dir (bool, optional) – This parameter is only relevant for undirected graphs. If True, it forces the method to write both edge directions in the file i.e. (src, dst) and (dst, src). If False, only one direction is stored. Default is True.
See also
-
split_alg¶ A string indicating the algorithm used to split edges in train and test sets.
-
split_id¶ An int used as an ID for this particular train/test split.
-
store_edgelists(train_path, test_path)[source]¶ Writes the sets of train and test node pairs to files with the specified names.
Parameters: - train_path (string) – Indicates the path where the train data will be stored.
- test_path (string) – Indicates the path where the test data will be stored.
-
test_edges¶ The set of test node pairs.
-
test_labels¶ A list of test node-pair labels. Labels can be either 0 or 1 and denote non-edges and edges, respectively (for SP they denote negative and positive links, respectively).
-
train_edges¶ The set of training node pairs.
-
train_frac¶ A float indicating the fraction of train edges out of all train and test edges.
-
train_labels¶ A list of train node-pair labels. Labels can be either 0 or 1 and denote non-edges and edges, respectively (for SP they denote negative and positive links, respectively).
-
-
class
evalne.evaluation.split.EvalSplit[source]¶ Bases:
evalne.evaluation.split.LPEvalSplitDeprecated and will be removed in v0.4.0. Use LPEvalSplit instead.
-
read_splits(filename, split_id, directed=False, nw_name='test', verbose=False)[source]¶ Reads the train and test edges and non-edges from files and initializes the class attributes.
Parameters: - filename (string) – The filename shared by all edge splits as given by the ‘store_train_test_splits’ method
- split_id (int) – The ID of the edge splits to read. As provided by the ‘store_train_test_splits’ method
- directed (bool, optional) – True if the splits correspond to a directed graph, false otherwise. Default is False.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is test.
- verbose (bool, optional) – If True print progress info. Default is False.
See also
evalne.utils.preprocess.read_train_test()- The low level function used for reading the sets of edges and non-edges.
evalne.utils.split_train_test.store_train_test_splits()- The files in the provided input path are expected to follow the naming convention of this function.
-
-
class
evalne.evaluation.split.LPEvalSplit[source]¶ Bases:
evalne.evaluation.split.BaseEvalSplitClass that provides a high level interface for managing/computing sets of train and test edges and non-edges for LP tasks. The class exposes the train edges and non-edges through the train_edges property and the test edges and non-edges through the test_edges property. Parameters used to compute these sets are also made available.
Notes
In link prediction the aim is to predict, given a set of node pairs, if they should be connected or not. This is generally solved as a binary classification task. For training the binary classifier, we sample a set of edges as well as a set of unconnected node pairs. We then compute the node-pair embeddings of this training data. We use the node-pair embeddings together with the corresponding labels (0 for non-edges and 1 for edges) to train the classifier. Finally, the performance is evaluated on the test data (the remaining edges not used in training plus another set of randomly selected non-edges).
-
compute_splits(G, nw_name='test', train_frac=0.51, split_alg='spanning_tree', owa=True, fe_ratio=1, split_id=0, verbose=False)[source]¶ Computes sets of train and test edges and non-edges according to the given input parameters and initializes the class attributes.
Parameters: - G (graph) – A NetworkX graph or digraph to compute the train test split from.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is ‘test’.
- train_frac (float, optional) – The proportion of train edges w.r.t. the total number of edges in the input graph (range (0.0, 1.0]). Default is 0.51.
- split_alg (string, optional) – A string indicating the algorithm to use for generating the train/test splits. Options are spanning_tree, random, naive, fast and timestamp. Default is spanning_tree.
- owa (bool, optional) – Encodes the belief that the network should respect or not the open world assumption. Default is True. If owa=True, train non-edges are sampled from the train graph only and can overlap with test edges. If owa=False, train non-edges are sampled from the full graph and cannot overlap with test edges.
- fe_ratio (float, optional) – The ratio of non-edges to edges to sample. For fr_ratio > 0 and < 1 less non-edges than edges will be generated. For fe_edges > 1 more non-edges than edges will be generated. Default 1, same amounts.
- split_id (int, optional) – The id to be assigned to the train/test splits generated. Default is 0.
- verbose (bool, optional) – If True print progress info. Default is False.
Returns: - train_E (set) – The set of train edges
- train_false_E (set) – The set of train non-edges
- test_E (set) – The set of test edges
- test_false_E (set) – The set of test non-edges
Raises: ValueError– If the edge split algorithm is unknown.
-
fe_ratio¶ A float indicating the ratio of non-edges to edges.
-
get_parameters()[source]¶ Returns the class properties except the sets of train and test node pairs, labels and train graph.
Returns: parameters – The parameters used when computing this split as a dictionary of parameters and values. Return type: dict
-
owa¶ A bool parameter indicating if the non-edges have been generated using the OWA (otherwise CWA).
-
set_splits(train_E, train_E_false=None, test_E=None, test_E_false=None, directed=False, nw_name='test', TG=None, split_id=0, split_alg='spanning_tree', owa=True, verbose=False)[source]¶ Sets the class attributes to the provided input values. The input train edges and non-edges as well as the test edges and non-edges are respectively joined to form the train_edges and test_edges class attributes. Train and test labels are also inferred from the input data.
Parameters: - train_E (set) – Set of train edges.
- train_E_false (set, optional) – Set of train non-edges. Default is None.
- test_E (set, optional) – Set of test edges. Default is None.
- test_E_false (set, optional) – Set of test non-edges. Default is None.
- directed (bool, optional) – True if the splits correspond to a directed graph, false otherwise. Default is False.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is test.
- TG (graph, optional) – A NetworkX graph or digraph containing all the train edges. If None, the graph will be generated from the set of train edges. Default is None.
- split_id (int, optional) – An ID that identifies this particular train/test split. Default is 0.
- split_alg (string, optional) – A string indicating the algorithm used to generate the train/test splits. Options are spanning_tree, random, naive, fast and timestamp. Default is spanning_tree.
- owa (bool, optional) – Encodes the belief that the network respects or not the open world assumption. Default is True. If owa=True, train non-edges are sampled from the train graph only and can overlap with test edges. If owa=False, train non-edges are sampled from the full graph and cannot overlap with test edges.
- verbose (bool, optional) – If True prints progress info. Default is False.
Raises: ValueError– If the train edge set is not provided.
-
-
class
evalne.evaluation.split.NREvalSplit[source]¶ Bases:
evalne.evaluation.split.BaseEvalSplitClass that provides a high level interface for managing/computing sets of train edges and non-edges for NR tasks. The class exposes the train edges and non-edges through the train_edges property. Test edges are not used for NR and therefore the test_edges property will be left empty. Parameters used to compute these sets are also made available.
Notes
In network reconstruction the aim is to asses how well an embedding method captures the structure of a given graph. The embedding methods are trained on a complete input graph. Hyperparameter tuning is performed directly on this graph (overfitting is, in this case, expected and desired). The embeddings obtained are used to perform link predictions and their quality is evaluated. Checking the link predictions for all node pairs is generally unfeasible, therefore a subset of all node pairs in the input graph are selected for evaluation.
-
compute_splits(G, nw_name='test', samp_frac=0.01, split_id=0, verbose=False)[source]¶ Computes sets of train edges and non-edges by randomly sampling elements from the adjacency matrix of G and initializes the class attributes.
Parameters: - G (graph) – A NetworkX graph or digraph to sample node pairs from.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is ‘test’.
- samp_frac (float, optional) – The fraction of node-pairs out of all possible ones to sample for NR evaluation. Default is 0.01 (1%).
- split_id (int, optional) – The id to be assigned to the train/test splits generated. Default is 0.
- verbose (bool, optional) – If True print progress info. Default is False.
Returns: - train_E (set) – The set of train edges.
- train_false_E (set) – The set of train non-edges.
Raises: ValueError– If the edge split algorithm is unknown.
-
get_parameters()[source]¶ Returns the class properties except the sets of train and test node pairs, labels and train graph.
Returns: parameters – The parameters used when computing this split as a dictionary of parameters and values. Return type: dict
-
samp_frac¶ A float indicating the fraction of node pairs out of all possible ones sampled for NR evaluation.
-
set_splits(TG, train_E, train_E_false=None, samp_frac=None, directed=False, nw_name='test', split_id=0, verbose=False)[source]¶ Sets the class attributes to the provided input values. The input train edges and non-edges are joined to form the train_edges class attribute. Train labels are also inferred from the input data.
Parameters: - TG (graph) – A NetworkX graph or digraph, the complete network from which train_E and train_E_false were sampled.
- train_E (set) – Set of train edges.
- train_E_false (set, optional) – Set of train non-edges. Default is None.
- samp_frac (float, optional) – The fraction of node-pairs out of all possible ones sampled for NR evaluation. Default is None.
- directed (bool, optional) – True if the splits correspond to a directed graph, false otherwise. Default is False.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is test.
- split_id (int, optional) – An ID that identifies this particular train/test split. Default is 0.
- verbose (bool, optional) – If True prints progress info. Default is False.
Raises: ValueError– If the train edge set is not provided.
-
-
class
evalne.evaluation.split.SPEvalSplit[source]¶ Bases:
evalne.evaluation.split.BaseEvalSplitClass that provides a high level interface for managing/computing sets of train and test positive and negative edges for SP tasks. The class exposes the train positive and negative edges through the train_edges property and the test positive and negative edges through the test_edges property. Parameters used to compute these sets are also made available.
Notes
In sign prediction the aim is to predict the sign (positive or negative) of given edges. The existence of the edges is assumed (i.e. we do not predict the sign of unconnected node pairs). Therefore, sign prediction is also a binary classification task similar to link prediction where, instead of predicting the existence of edges or not, we predict the signs for edges we know exist. Unlike for link prediction, in this case we do not need to perform negative sampling, since we already have both classes (the positively and the negatively connected node pairs).
-
compute_splits(G, nw_name='test', train_frac=0.51, split_alg='spanning_tree', split_id=0, verbose=False)[source]¶ Computes sets of train and test positive and negative edges according to the given input parameters and initializes the class attributes.
Parameters: - G (graph) – A NetworkX graph or digraph to compute the train test split from.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is ‘test’.
- train_frac (float, optional) – The proportion of train edges w.r.t. the total number of edges in the input graph (range (0.0, 1.0]). Default is 0.51.
- split_alg (string, optional) – A string indicating the algorithm to use for generating the train/test splits. Options are spanning_tree, random, naive, fast and timestamp. Default is spanning_tree.
- split_id (int, optional) – The id to be assigned to the train/test splits generated. Default is 0.
- verbose (bool, optional) – If True print progress info. Default is False.
Returns: - train_E (set) – The set of train positive edges.
- train_false_E (set) – The set of train negative edges.
- test_E (set) – The set of test positive edges.
- test_false_E (set) – The set of test negative edges.
Raises: ValueError– If the edge split algorithm is unknown.
-
set_splits(train_E, train_E_false=None, test_E=None, test_E_false=None, directed=False, nw_name='test', TG=None, split_id=0, split_alg='spanning_tree', verbose=False)[source]¶ Sets the class attributes to the provided input values. The input train positive and negative edges as well as the test positive and negative edges are respectively joined to form the train_edges and test_edges class attributes. Train and test labels (0 or 1 representing negative and positive edges, respectively) are also inferred from the input data.
Parameters: - train_E (set) – Set of positive train edges.
- train_E_false (set, optional) – Set of negative train edges. Default is None.
- test_E (set, optional) – Set of positive test edges. Default is None.
- test_E_false (set, optional) – Set of negative test edges. Default is None.
- directed (bool, optional) – True if the splits correspond to a directed graph, false otherwise. Default is False.
- nw_name (string, optional) – A string indicating the name of the dataset from which this split was generated. This is required in order to keep track of the evaluation results. Default is test.
- TG (graph, optional) – A NetworkX graph or digraph containing all the train edges (positive and negative). If None, the graph will be generated from the sets of positive and negative train edges. Default is None.
- split_id (int, optional) – An ID that identifies this particular train/test split. Default is 0.
- split_alg (string, optional) – A string indicating the algorithm used to generate the train/test splits. Options are spanning_tree, random, naive, fast and timestamp. Default is spanning_tree.
- verbose (bool, optional) – If True prints progress info. Default is False.
Raises: ValueError– If the train edge set is not provided.
-